Clockor2: Inferring global and local strict molecular clocks using root-to-tip regression
Our webapp for root-to-tip regression Clockor2 (clockor2.github.io) has just been published in Systematic Biology .
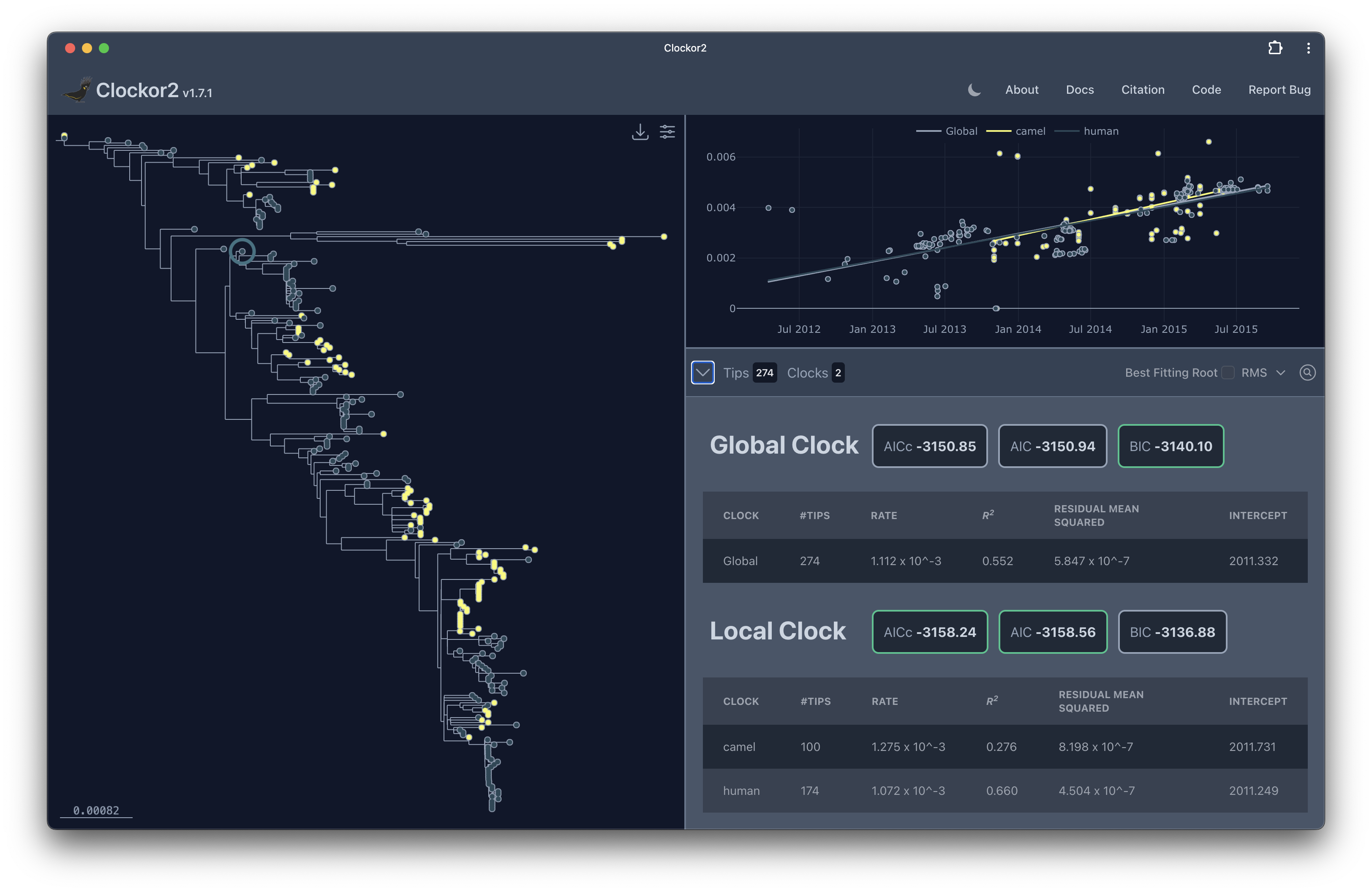
Clockor2 is a webapp for performing root-to-tip regression on phylogenetic trees. Root-to-tip regression is a method for estimating the rate of molecular evolution in a phylogenetic tree. It’s a simple method that can be used to calibrate strict molecular clocks. The method is based on the assumption of clock-like evolution, that mutations accumulate at some fixed rate. Root-to-tip regression is widely used in phylogenetics and is a key quality control step in many phylogenetic analyses. A special feature of Clockor2 is that it allows you to fit multiple clocks in different parts of the tree (a clock or 2).
Clockor2 is one of many modern tools that are moving computation back to the browser. The web ecosystem as come a long way since the days of running a janky script on a server to obtain the reverse complement of your DNA sequence. Modern features like WebAssembly, WebWorkers, and WebGL have made it possible to run complex analyses directly in the browser.
Clockor2 takes advantage of modern web technologies to provide a fast and responsive user interface. The tree viewer is powered by phylocanvas.gl, a WebGL-based phylogenetic tree viewer. WebGL allows us to render large trees with thousands of tips in the browser by offloading the rendering to the GPU. The library underlying phylocanvas.gl is Deck.gl which is also used by tools like taxonium.org. The regression plotting is powered by Plotly.js, a powerful charting library that allows us to create interactive plots, which also uses WebGL for plotting large datasets.
The most computationally intensive part of Clockor2 is finding the best fitting root. This involves iteratively re-rooting the tree and calculating the coefficient of determination (R^2^) or residual mean square (RMS) for each root. To speed up this process we use WebWorkers to parallelize the computation (potential rooting locations are divided into the number of cores available). WebWorkers allow us to run the computation in the background without blocking the main thread. This means that the user can continue to interact with the webapp while the computation is running. This has the added benefit that the computation will run faster on multi-core machines.
Clockor2 is open source and available on GitHub. Github also serves as our deployment platform. We use GitHub Actions to automatically build and deploy the webapp to GitHub Pages whenever we push a new release. This means that the webapp is always up-to-date with the latest features and bug fixes. We also use GitHub Issues to track bugs and feature requests. If you have any issues or feature requests, please feel free to open an issue on GitHub.
We hope that Clockor2 will be a useful tool for the phylogenetics community. We are always looking for feedback and feature requests. If you have any questions or comments, please feel free to reach out to us on GitHub.
If you use Clockor2 in your research, please cite our paper:
Leo A Featherstone, Andrew Rambaut, Sebastian Duchene, Wytamma Wirth, Clockor2: Inferring global and local strict molecular clocks using root-to-tip regression, Systematic Biology, 2024;, syae003, https://doi.org/10.1093/sysbio/syae003


Comments